Myeloid malignancies are clonal disorders of haematopoietic origin characterized by excessive proliferation, abnormal self-renewal, and/or differentiation defects of haematopoietic stem cells (HSCs) and myeloid progenitor cells¹. These malignancies consist mainly of myeloproliferative neoplasms (MPNs), myelodysplastic syndrome (MDS), and acute myeloid leukemia (AML). Malignant myeloid cells tend to be genotypically and phenotypically heterogeneous with multiple subclones. Genetic heterogeneity, in particular, may present an obstacle that can impact targeted treatment efficacy.
Malignancies in myeloid cells arise from genetic and epigenetic alterations in several key genes that encode proteins belonging to five main classes, namely:
This blog, the first of five in a series, is intended to shed light on one key gene in each of the above mentioned classes, respectively. For the first class, signaling pathway proteins, we will discuss FLT3 gene. Specifically, we will discuss FLT3’s normal function, its implications in myeloid malignancies, and the role of next-generation sequencing (NGS) in genetic identification and disease management of patients with FLT3 genetic alterations.
Fms-like tyrosine kinase 3 or FLT3 gene, located on chromosome 13, encodes a receptor tyrosine kinase which plays a pivotal role in regulating normal haematopoiesis. Upon activation, this receptor phosphorylates and activates multiple cytoplasmic effector molecules in pathways involved in apoptosis, proliferation, and differentiation of haematopoietic cells in the bone marrow³. Genetic alterations in the FLT3 gene may induce constitutive activation of this tyrosine kinase receptor and enhance excessive cell proliferation, leading to AML and acute lymphoblastic leukemia (ALL).
FLT3 gene alterations are common and recurring mutations in myeloid malignancies in AML patients. It is estimated that 30% of newly diagnosed AML patients harbour mutations in the FLT3 gene.
Major mutations in the FLT3 gene are structural variants (SVs) such as internal tandem duplications (FLT3-ITD) and tyrosine kinase domain (FLT3-TKD). FLT3-ITD mutations usually confer a poor prognosis at diagnosis with an increased risk for relapse in AML. According to a recent article by Daver et al., the prognostic impact of FLT3-ITD mutations is influenced by different factors, including allele ratio (AR), insertion site, ITD length, co-mutation with NPM1, and karyotype⁴. FLT3-TKD mutations are also associated with poor prognosis, specifically those point mutations involving codon D835 or I836; however, the prognostic outcome of these mutations remains speculative and unclear compared to FLT3- ITDs. Additionally, structural variants in FLT3 are a target for therapeutic agents that may assist in disease management.
Polymerase chain reaction (PCR) is the current standard method utilised in clinical laboratories to detect FLT3-ITDs. PCR is desirable for its short TAT and high sensitivity, especially in monitoring minimal residual disease (MRD) in AML.
Nevertheless, as the number of clinically relevant somatic AML mutations increases, it is now increasingly attractive to incorporate FLT3-ITD testing into multiplex assays to identify many relevant somatic mutations simultaneously, using next-generation sequencing (NGS)⁵. Utilising a well-designed NGS panel with a high coverage may offer higher sensitivity than PCR in detecting FLT3-ITDs, allowing laboratories to incorporate FLT3-ITD and other variants into their in-house NGS panel.
Additionally, NGS is a better technique than PCR in detecting a wide range of mutational shifts and disease progression from one malignancy to another after initial diagnosis. Hence, providing patients with better-tailored treatment through the timely use of targeted therapies at relapse.
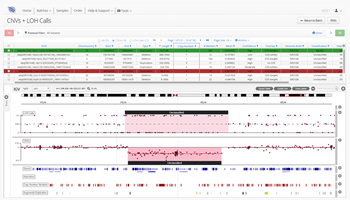
As one of the first steps in many NGS data analysis pipelines, accurate variant calling is often critical to downstream analysis and interpretation. Here, we take a look at variant calling best practice through a modern lens.
Read
A high-quality sequencing library is the linchpin to generating good sequencing data. We discuss our six top tips to help you improve your sequencing library.
Read
We discuss the development and current state of sequencing technologies, and where the future of NGS may take us...
Read
FISH is a cytogenetic technique utilised in labs to detect chromosomal abnormalities in both cancer and constitutional specimens. In this blog learn about the advantages of FISH...
Read
While liquid biopsy may present an attractive alternative to a solid biopsy, it also has limitations. Here, we shed light on some advantages, limitations, and future outlook for liquid biopsy in oncology clinical practice.
Read
Find out about the benefits and differentiating factors of the three most commonly used NGS technologies; targeted gene panels, whole-exome sequencing (WES) and whole-genome sequencing (WGS).
Read
Next generation sequencing (NGS) is now in routine use for a broad range of research and clinical applications. Facilitating the detection of a wide variety of mutations, focus has never been higher on the value of making the correct choice for the initial sequence enrichment step, which, if poorly designed, can be a source of bias and error in the downstream sequencing assay.
Read